SIR model¶
In [SZ14] the following SIR model is considered
where \(A>0\) is the recruitment rate of susceptible population; \(d>0\) is the per capita natural death rate of the population; \(\nu>0\) is the per capita disease-induced death rate; \(\mu>0\) is the per capita recovery rate of infectious individual. The funtion \(\mu\) is set to
Overview¶
In this demo we will
Compute a curve of equilibria, parametrized by \(b\).
Detect a Hopf and limit point.
Start continuation form the Hopf point in two parameters \((\mu_1, b)\).
Detect a Bogdanov-Takens points.
Start continuation form the Bogdanov-Takens points in two parameters \((\mu_1, b)\).
Compare the predicted and computed homoclinic bifurcation curve emanating from the Bogdanov-Takens point in parameters space.
Compare a range of predictors for the homoclinic solutions emanating from the Bogdanov-Takens point with the corrected homoclinic solutions curve in phase-space.
Create a convergence plot comparing the different homoclinic approximations derived in [Kuz21].
Load MatCont¶
Before we can start using MatCont we need to add the main directory of
MatCont, as well as various subdirectories of MatCont, to the MATLAB
search path. This is done in the code below. The variable matcont_home
should point to the main directory of MatCont.
clear all
matcontpath = '../';
addpath(matcontpath)
addpath([matcontpath, 'Equilibrium'])
addpath([matcontpath, 'Systems'])
addpath([matcontpath, 'Hopf'])
addpath([matcontpath, 'Homoclinic'])
addpath([matcontpath, 'LimitPoint'])
addpath([matcontpath, 'LimitCycle'])
addpath([matcontpath, 'Continuer'])
addpath([matcontpath, 'MultilinearForms'])
addpath([matcontpath, 'Utilities'])
set(groot, 'defaultTextInterpreter', 'LaTeX');
set(0,'defaultAxesFontSize',15)
Set the odefile¶
Next we set the variable odefile to the system file previously generated by
the notebook SIRmodel.ipynb.
odefile=@SIRmodel;
Define equilibrium¶
As in [SZ14] we fix the parameters \(A = 20, \mu_0 = 10, d = 1/10, \nu = 1,\) and \(\beta = 11.5\).
A = 20;
mu0 = 10;
d = 1/10;
nu = 1;
beta = 11.5;
For any endemic equilibrium \(E(S, I, R)\), its coordinates satisfy
mu = @(b,mu1,I) mu0 + (mu1-mu0)*b/(I+b);
S = @(b,I,mu1) (A-(d+nu+mu(b, mu1, I))*I)/d;
R = @(b,I,mu1) mu(b, mu1, I)*I/d;
and the coordinate \(I\) should be the positive root of the quadratic equation
where
R0 = @(mu1) beta/(d+nu+mu1);
Ascr = (d+nu+mu0)*(beta-nu);
Bscr = @(b,mu1) (d+nu+mu0-beta)*A+(beta-nu)*(d+nu+mu1)*b;
Cscr = @(b,mu1) (d+nu+mu1)*A*b*(1-R0(mu1));
with
the basic reproduction number. The equation \(f(I)=0\) may have two roots if \(\Delta_{0}>0\) which are denoted as
delta0 = d+nu+mu0;
delta1 = @(mu1) d+nu+mu1;
Delta0 = @(b,mu1) (beta-nu)^2*delta1(mu1)^2*b^2-2*A*(beta-nu)*(beta*(mu1-mu0)+delta0*(delta1(mu1)-beta))*b+A^2*(beta-delta0)^2;
I1 = @(b,mu1)(-Bscr(b,mu1)-sqrt(Delta0(b,mu1)))/(2*Ascr);
I2 = @(b,mu1)(-Bscr(b,mu1)+sqrt(Delta0(b,mu1)))/(2*Ascr);
By setting \(b=0.022\) and \(\mu_1 = 10.45\) we are slightly below the Hopf curve in Figure 4 of [SZ14].
b=0.022;
mu1=10.45;
p(1) = mu1;
p(2) = b;
x = [S(b,I2(b,mu1),mu1); I2(b,mu1); R(b,I2(b,mu1),mu1)];
To refer to the parameters throughout the script we create a cell array of
strings containing the parameter names. This is then converted into a
struct. This allows us to refer to the parameters as ind.parametername,
similar as done in DDE-BifTool.
parnames = {'mu1','b'};
cind = [parnames;num2cell(1:length(parnames))];
ind = struct(cind{:});
ap = [ind.mu1, ind.b]; % continuation parameters
Continue equilibrium in parameter \(b\)¶
To continue the equilibrium in parameter \(b\), we first need to obtain a
tangent vector to the curve. This is done by the function init_EP_EP. Then we
use the function contset to obtain a struct containing a list of options
which is passed on to the continuer. By adjusting the values of the fields of
the opt struct we set the minimum, initial and maximum set sizes, as well
as the maximum number of points to continue and weather or not to detect
bifurcation points (opt.Singularities) on the equilibrium curve. We also
increase the accuracy for locating detected bifurcations (TestTolerance) and
the maximum number of iterations that may be used to achieve this
(MaxTestIters). This improves the homoclinic predictor which depend directly
on the accuracy of the located Bogdanov-Takens point.
Finally, we continue the curve using the function cont.
[x1_pred, v1_pred] = init_EP_EP(odefile, x, p, ind.b);
opt = contset;
opt.MaxNumPoints = 1000;
opt.Singularities = 1;
opt.TestTolerance = 1e-15;
opt.MaxTestIters = 10;
opt.MinStepsize = 0.001;
[eqbr_x, ~, eqbr_bif_data] = cont(@equilibrium, x1_pred, v1_pred, opt);
first point found
tangent vector to first point found
label = H , x = ( 195.380284 0.041037 4.168310 0.022089 )
First Lyapunov coefficient = 3.908102e-04
label = LP, x = ( 198.069839 0.016933 1.743895 0.033419 )
a=-2.413445e+00
elapsed time = 0.7 secs
npoints curve = 1000
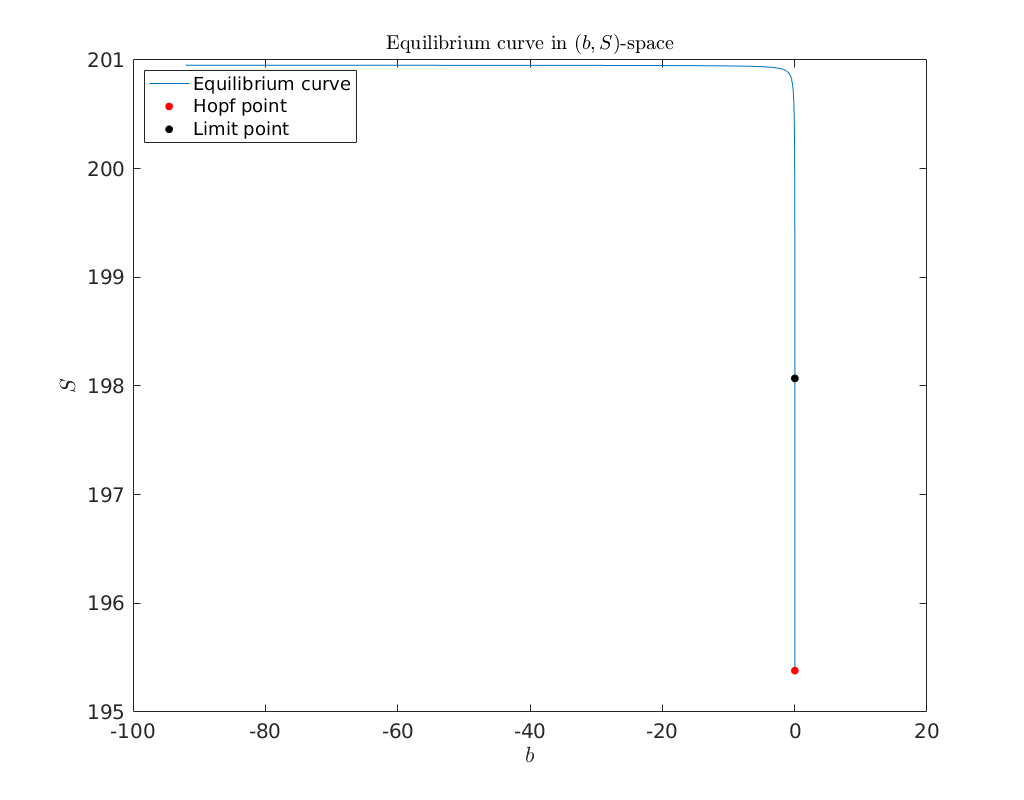
There is one limit point detected (LP and one Hopf bifurcation point (H). The
array struct eqbr_bif_data contains information about the detected
bifurcation points. We use this to extract the index of the detected bifurcation
points on the equilibrium curve eqbr_x. The equilibrium curve eqbr_x is
just a two dimensional array. Each column consists of a point on the curve.
The frist three rows contain the point \((S,I,R)\) while the last row contains
the parameter \(b\).
Below we plot the equilibrium curve eqbr_x, together with the detect Hopf and
limit point, in \((b,S)\)-space.
%plot --width 1024 --height 800
hopfPointInfo = eqbr_bif_data(strcmp({eqbr_bif_data.msg}, 'Hopf')==1);
limitPointsInfo = eqbr_bif_data(strcmp({eqbr_bif_data.msg}, 'Limit point')==1);
HopfPoint1 = eqbr_x(:,hopfPointInfo.index);
limitpoint1 = eqbr_x(:,limitPointsInfo.index);
plot(eqbr_x(4,:), eqbr_x(1,:)); hold on
plot(HopfPoint1(4), HopfPoint1(1), '.r' ,'MarkerSize', 20)
plot(limitpoint1(4), limitpoint1(1), '.k' ,'MarkerSize', 20)
xlabel('$b$')
ylabel('$S$')
legend({'Equilibrium curve', 'Hopf point', 'Limit point'}, 'Location', 'NorthWest')
title('Equilibrium curve in $(b,S)$-space', 'fontsize', 15)
Continue Hopf point¶
We continue the Hopf point using again the function cont. We use the same
continuation options as defined above in the struct opt.
hopf1.par = [mu1, b];
hopf1.par(ind.b) = eqbr_x(4, hopfPointInfo(1).index);
hopf1.x = eqbr_x(1:3, hopfPointInfo(1).index);
hopf1.x = x;
[hopf_x, hopf_v] = init_H_H(odefile, hopf1.x, hopf1.par', ap);
opt.MaxNumPoints = 500;
opt.MaxStepsize = .01;
opt.Singularities = 1;
[hopf_br, ~, hopf_br_bif] = cont(@hopf, hopf_x, hopf_v, opt);
first point found
tangent vector to first point found
label = GH, x = ( 195.599878 0.039046 3.970613 10.427886 0.025488 0.012691 )
l2=-8.624030e-05
Warning: Matrix is close to singular or badly scaled. Results may be inaccurate. RCOND = 1.423059e-17.
> In nf_H (line 27)
In hopf>testf (line 150)
In cont>EvalTestFunc (line 810)
In cont (line 506)
Warning: Matrix is close to singular or badly scaled. Results may be inaccurate. RCOND = 4.847349e-18.
> In nf_H (line 28)
In hopf>testf (line 150)
In cont>EvalTestFunc (line 810)
In cont (line 506)
label = BT, x = ( 198.059382 0.017026 1.753334 10.450822 0.033239 0.000000 )
(a,b)=(-1.298117e-03, 1.219168e-02)
elapsed time = 1.3 secs
npoints curve = 500
Continue homoclinic curve emanating from the Bogdanov-Takens point¶
To continue the homoclinic curve emanating from the Bogdanov-Takens point we
use the functions BT_Hom_set_options and init_BT_Hom to obtain an initial
approximation to the homoclinic solution (hom_x) as well as a tangent vector
to the discretized homoclinic solution (hom_v) as described in
Initial prediction of homoclinic orbit near Bogdanov-Takens point 1.
bt_points_info = hopf_br_bif(strcmp({hopf_br_bif.label}, 'BT')==1);
bt.x = hopf_br(1:3, bt_points_info.index);
bt.par = hopf_br(4:5, bt_points_info.index);
BToptions = BT_Hom_set_options();
[hom_x, hom_v] = init_BT_Hom(odefile, bt, ap, BToptions);
opt = contset;
opt.Singularities = 0;
homoclinic_br = cont(@homoclinic, hom_x, hom_v, opt);
Center manifold coefficients' accuracy: 7.460699e-13
BT normal form coefficients:
a=-1.298117e-03, b=1.219168e-02
The initial perturbation parameter epsilon: 1.000000e-01
The initial amplitude: 0.524008
The initial half-return time T: 358.423
The initial distance eps0: 0.00239581
The initial distance eps1: 0.000941365
first point found
tangent vector to first point found
elapsed time = 18.7 secs
npoints curve = 300
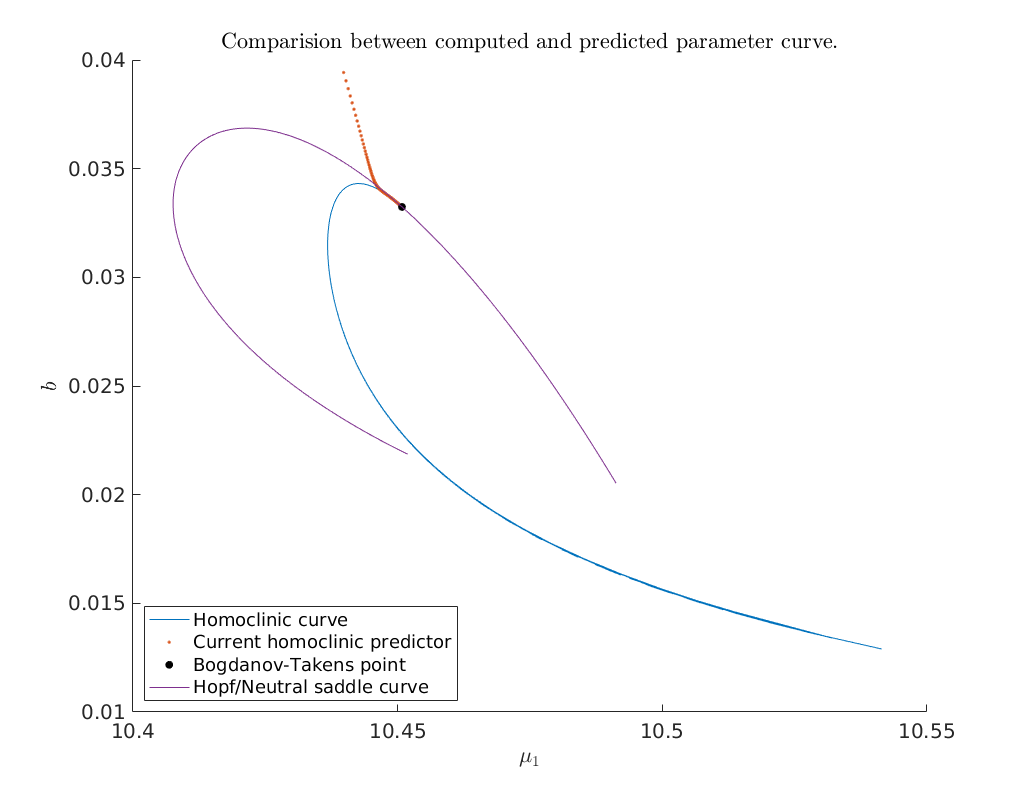
Compare predicted with computed parameters¶
Now that we have obtained a curve of homoclinic orbits (homoclinic_br) we
compare the computed curve in parameter space with the predicted curve we
construct below. To do so, we use the function BT_nmfm_orbital to obtain the
smooth orbital normal form coefficients, i.e. \(a\) and \(b\), and the coefficients
of the transformation \(K\) between the parameters of the system and the parameters
of the smooth orbital normal form on the center manifold, see
[Kuz21]. We also plot the Hopf curve which can be compared with
Figure 4 in [SZ14].
global homds
hold on
% plot computed homoclinic parameter curve
plot(homoclinic_br(homds.PeriodIdx+1,:), ...
homoclinic_br(homds.PeriodIdx+2,:));
% Bogdanov-Takens parameter-dependent smooth orbital normal form coefficients
bt = BT_nmfm_orbital(odefile, bt, ap, BToptions);
a = bt.nmfm.a;
b = bt.nmfm.b;
K10 = bt.nmfm.K10;
K01 = bt.nmfm.K01;
K02 = bt.nmfm.K02;
K11 = bt.nmfm.K11;
K03 = bt.nmfm.K03;
% construct predictor as in the paper
eps = linspace(0, 0.3);
beta1 = -4*a^3/b^4*eps.^4;
tau0 = 10/7;
tau2 = 288/2401;
beta2 = a/b*(tau0 + tau2*eps.^2).*eps.^2;
alpha = K10.*beta1 + K01.*beta2 + 1/2*K02.*beta2.^2 ...
+ K11.*beta1.*beta2 + 1/6*K03.*beta2.^3;
alpha = bt.par(ap) + alpha;
% plot currect predictor
plot(alpha(1,:), alpha(2,:), '.')
% plot Bogdanov-Takens point
plot(bt.par(ind.mu1), bt.par(ind.b), '.k', 'MarkerSize', 20)
plot(hopf_br(4,:), hopf_br(5,:))
% set axis labels and legend
xlabel('$\mu_1$')
ylabel('$b$')
legend({'Homoclinic curve', 'Current homoclinic predictor', ...
'Bogdanov-Takens point', 'Hopf/Neutral saddle curve'}, 'Location', 'SouthWest')
title('Comparision between computed and predicted parameter curve.')
Center manifold coefficients' accuracy: 7.460699e-13
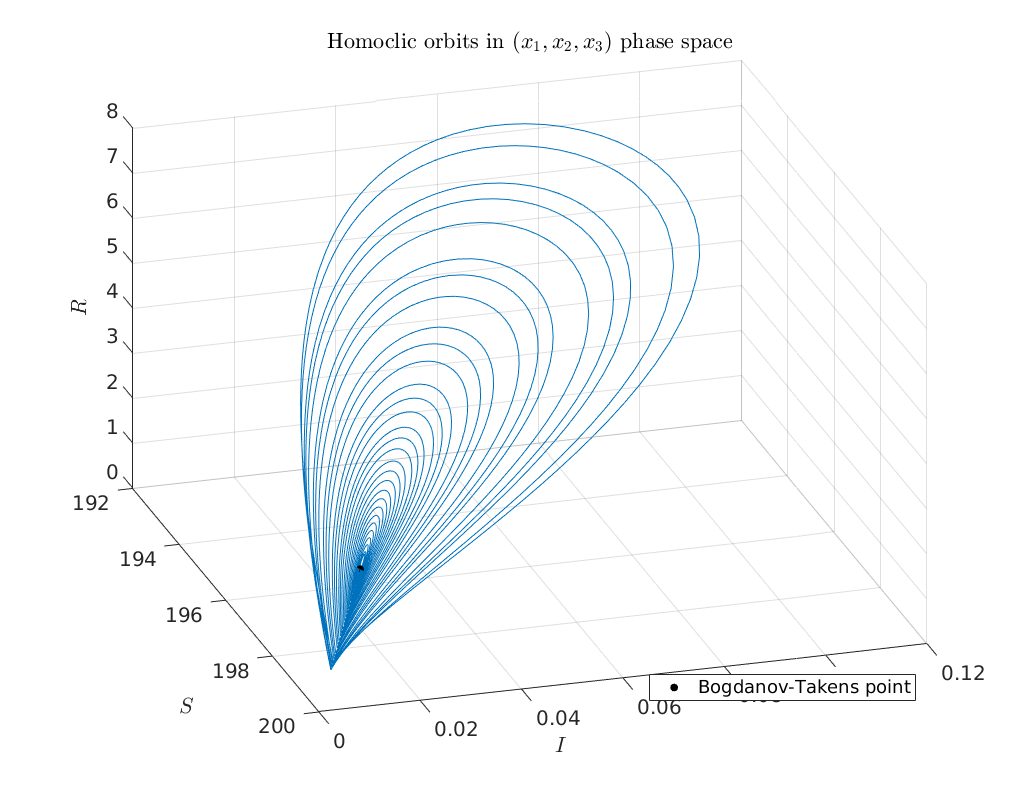
Plot of continued homoclinic solutions in phase-space¶
To obtain an impression of the homoclinic solutions we plot the computed homoclinic orbits in \((S,I,R)\) phase-space.
hold on
plot3(homoclinic_br(homds.coords(1:homds.nphase:end), 1:10:end), ...
homoclinic_br(homds.coords(2:homds.nphase:end), 1:10:end), ...
homoclinic_br(homds.coords(3:homds.nphase:end), 1:10:end), ...
'Color', [0 0.4470 0.7410], 'HandleVisibility', 'Off')
xlabel('$S$')
ylabel('$I$')
zlabel('$R$')
plot3(bt.x(1), bt.x(2), bt.x(3), '.k' ,'MarkerSize', 20)
legend('Bogdanov-Takens point', 'Location', 'SouthEast')
title('Homoclic orbits in $(x_1,x_2,x_3)$ phase space')
grid on
view([73, 33]);
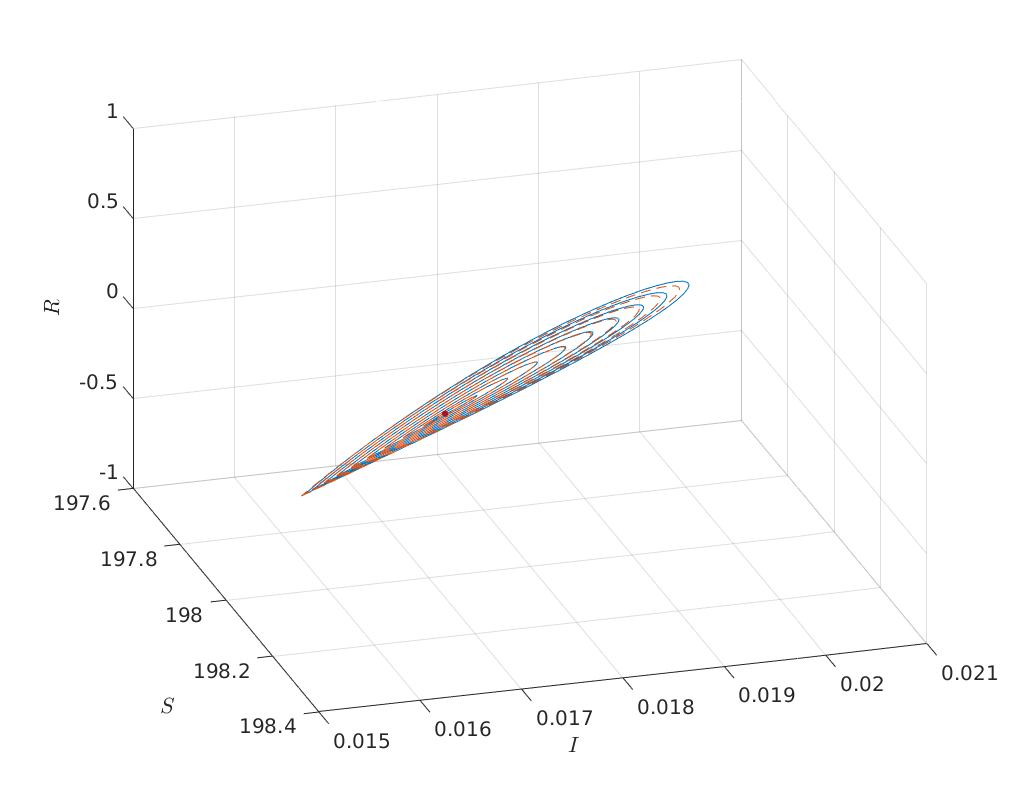
Predictors of orbits for various epsilons¶
Below we compute for a large range of amplitudes the predicted and corrected
homoclinic solutions and compare them in phase space. We see that even with an
amplitude of 1 the predicted homoclinic orbit is still relatively close.
options = BT_Hom_set_options();
options.messages = false;
options.correct = false;
options.TTolerance = 1.0e-05;
amplitudes = linspace(1.0e-03, 1,10);
XPredicted = zeros(494,length(amplitudes));
XCorrected = zeros(494,length(amplitudes));
for j=1:length(amplitudes)
options.amplitude = amplitudes(j);
[x_pred, v0] = init_BT_Hom(odefile, bt, ap, options);
XPredicted(:,j) = x_pred;
try
XCorrected(:,j) = newtcorr(x_pred, v0);
catch
warning('Didn''t convergence to homoclinic solution')
end
end
hold on
cm = lines;
plot(XPredicted(homds.coords(1:homds.nphase:end),1:10), ...
XPredicted(homds.coords(2:homds.nphase:end),1:10), ...
'color', cm(1,:))
plot(XCorrected(homds.coords(1:homds.nphase:end),1:10), ...
XCorrected(homds.coords(2:homds.nphase:end),1:10), ...
'--', 'color', cm(2,:))
plot(bt.x(1), bt.x(2), '.', 'MarkerSize', 16)
xlabel('$S$')
ylabel('$I$')
zlabel('$R$')
grid on
view([73, 33]);
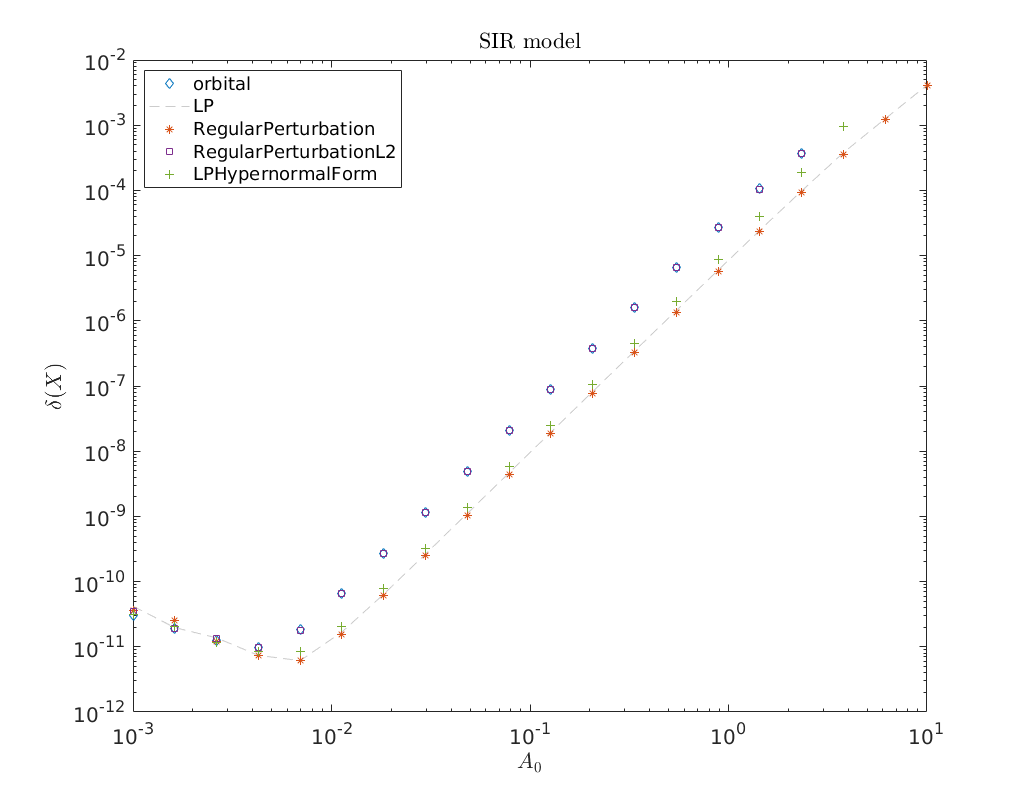
Convergence plot¶
We finish this notebook with a log-log convergence plot comparing the different
third order homoclinic approximation methods derived in [Kuz21]
to approximate the homoclinic solutions near the first Bogdanov-Takens point.
On the abscissa is the amplitude \(A_0\) and on the ordinate the relative error
\(\delta\) between the constructed solution (x_pred) to the defining system for the
homoclinic orbit and the Newton corrected solution (x_corrected).
BToptions = BT_Hom_set_options();
BToptions.TTolerance = 1e-05;
BToptions.messages = false;
BToptions.correct = false;
amplitudes = logspace(-3, 1, 20);
methodList = {'orbital', 'LP', 'RegularPerturbation', ...
'RegularPerturbationL2', 'LPHypernormalForm'};
relativeErrors = {};
for i=1:length(methodList)
BToptions.method = methodList{i};
relativeErrors{i} = zeros(size(amplitudes));
for j=1:length(amplitudes)
BToptions.amplitude = amplitudes(j);
[x_pred, v0] = init_BT_Hom(odefile, bt, ap, BToptions);
try
x_corrected = newtcorr(x_pred, v0);
relativeErrors{i}(j) = norm(x_corrected-x_pred)/norm(x_corrected);
catch
warning('Did not converge.')
continue
end
end
end
cm = lines();
loglog(amplitudes, relativeErrors{1}(:), 'd', ...
amplitudes, relativeErrors{2}(:), '--', ...
amplitudes, relativeErrors{3}(:), '*', ...
amplitudes, relativeErrors{4}(:), 's', ...
amplitudes, relativeErrors{5}(:), '+')
legend(methodList, 'Location', 'NorthWest')
title('SIR model')
xlabel('$A_0$')
ylabel('$\delta(X)$')
ax = gca;
ax.ColorOrder = [cm(1,:); [0.8 0.8 0.8]; cm(2,:); cm(4,:); cm(5,:)];
Warning: Did not converge.
Warning: Did not converge.
Warning: Did not converge.
Warning: Did not converge.
Warning: Did not converge.
Warning: Did not converge.
Warning: Did not converge.
Warning: Did not converge.